One of our points of pride for being able to pool, standardize, and share gene and variant annotation data as a service, is that our service is fast! The reason that MyGene.info and MyVariant.info are made with speed in mind is because we want them to be useful to bioinformaticians and tool/resource developers alike! How can we tell if we’ve successfully provided a useful service?
One measure we LOVE, is when a user builds something useful or amazing with our service--and some amazing researchers at Washington University in St. Louis have done just that! Without further ado, we’d like to introduce CIViC.
In one tweet or less, introduce us to CIViC: CIViC is an open access, open source, community-driven web resource for Clinical Interpretation of Variants in Cancer. Our goal is to enable precision medicine by providing an educational forum for dissemination of knowledge and active discussion of the clinical significance of cancer genome alterations.
How does CIViC use MyGene.info or MyVariant.info services? CIViC has integrated MyGene.info into the interface for all available genes. We show users the gene name, symbol, relevant aliases, coordinate information, protein domains, and associated pathways. We subsequently link each gene to MyGene.info details for more information. An example of how we integrated MyGene.info into the CIViC interface is shown below:
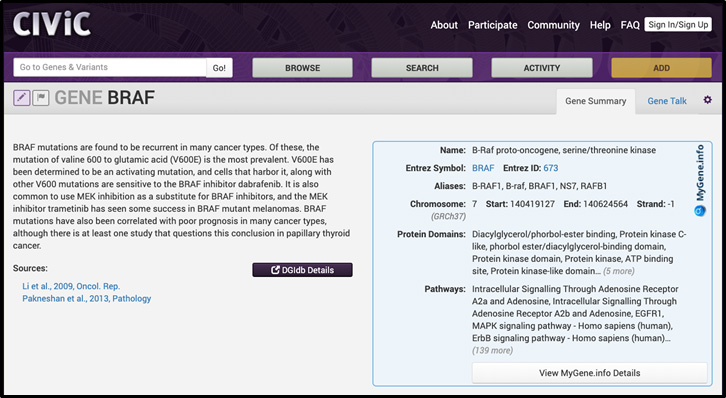
CIViC has also integrated MyVariant.info into the interface for all eligible variants. We show users the ClinVar ID, Clinical Significance, COSMIC ID, dbSNP RSID, HGVS ID, SnpEff Effect, SnpEff Impact and gnomAD frequency. We subsequently link each variant to MyVariant.info details for much more information. An example of how we integrated MyVariant.info into the CIViC interface is shown below:
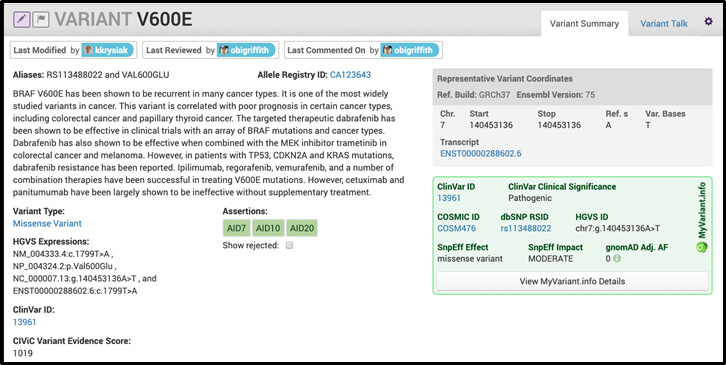
As a resource on gene variants in cancer, CIViC stands out for a number of reasons. First, CIViC is open source (under a CC0 1.0 Universal license). This alone is not unusual in bioinformatics, however, CIViC is also crowdsourced and community driven. How big is the contributor pool to CIViC and in what ways has using crowdsourcing shaped the evolution of the resource? Currently, CIViC has 175 unique contributors to the interface, 80% of which are outside the McDonnell Genome Institute organization. These other organizations include the Charité Comprehensive Cancer Center, the University Health Network, the Clinical Genome Resource (ClinGen), the BC Cancer Center’s Personalized Onco-Genomics (POG) program, the Treehouse Childhood Cancer Initiative and others. Additionally, CIViC has received large contributions from commercial entities including over 1,700 evidence statements from Illumina in 2017. Crowdsourcing has shaped the evolution of the resource in many ways. Specifically, we are able to increase total amount of curation, increase dedicated curation, and reduce bias associated with variant identification and curation. For example, the University Health Network in Toronto has dedicated an extraordinary amount of time to extensively curate VHL variant evidence in the database. To date, this has resulted in over 624 variants, 1527 evidence items and 4 assertions for VHL alone. We have used crowdsourcing curation to harness the knowledge of domain experts into a single database that is easily accessible by the community.
Another unique feature of CIViC is that the data is publically available for all users. I remember reading a statement about open data licensing and marveling at the speed with which CIViC made the transition towards public domain licensing. Was the data licensing issue something that was discussed or controversial within the CIViC community, or was the CIViC community supportive of open data from the get go?
The CIViC database has always believed in a policy of open-source software development and open-access for public data use. This was a conscious decision that CIViC made to overcome challenges of siloed data that currently exist in academia and industry. These policies serve as the foundation for the database and create a unique tool that can help to advance the genomics field. In the early days, we initially considered a CC-BY license but after reviewing community discussions on the benefits of public domain attribution we adopted CC0.
One criticism of crowdsourcing and open data seems to be concern about data quality when anyone is allowed to contribute. CIViC seems to handle this using moderation, but as well as using an evidence-based statement model similar to what you’d see on Wikidata. Was CIViC inspired by Wikidata at all in the use of this model? Does CIViC import its data into Wikidata as part of its open data efforts? CIViC was indeed inspired by the Wikidata model in terms of being open to the public. We also were inspired by Wikipedia “Talk” pages. All curated content in CIViC tracks comments and revisions in a corresponding talk page. We recognize that data quality is excruciatingly important, especially when it comes to annotation of somatic variants for patients. Therefore, items entered into the database have a process for internal review. For example, when an evidence item is added to the database it is not visible (by default), and it is not yet accepted. Editors or other curators review the submission and can optionally propose changes to the evidence statement. Ultimately, only editors can accept or reject the evidence statement or proposed changes. Changes and new submissions cannot be accepted by the same individual who submitted them. To further ensure that items within CIViC are correct, if any individual suspects an issue with an evidence item, the statement can be flagged for subsequent review. Additionally, there are requirements for becoming an editor. Specifically, editors must have attained a sufficient degree of education, they must be extensively familiar with the CIViC interface, have a demonstrated track record of successful curation within the database, and must be approved by at least two existing editorial members. More information can be found on the CIViC help docs on how to become a contributor and editor for the CIViC team.
One impressive feature of CIViC is how it has become an integral part of the open data loop. Although CIViC utilizes MyGene.info and MyVariant.info to consume data, the CIViC community also curates data and feeds that curated data into open data services like MyVariant.info. Was it difficult for CIViC to make the leap from data consumer to data distributor? Data distribution has been a goal since the origin of the CIViC database. Therefore, throughout development of CIViC, we have strived to create infrastructure to allow for easy data sharing in the hopes that other organizations would incorporate CIViC data and features into their research or clinical workflows. All data are available through nightly releases and our open/documented API. Furthermore, much effort by the CIViC team has focused on integrating CIViC into various interfaces including: cBioPortal, Allele Registry, CRAVAT and many others. We are so appreciative of CIViC’s amazing team of developers, students, collaborators, and scientists who have worked countless hours to distribute CIViC data through these channels.
As a community-driven project, how does CIViC recruit and retain its contributors? Are there citizen scientists in the CIViC community or does the CIViC community primarily consist of domain experts? CIViC has a variety of different curators who contribute to the database in varied ways. We have students with a range of education, who have continued to contribute even after they leave the lab. These individuals can curate variant coordinates, add evidence items, and moderate variant information. Additionally, we have cancer genomic groups whom mostly consume information, but also contribute evidence items or assertions when they encounter academic data that has not yet been curated within CIViC. Some of our most significant contributors are individuals who discovered CIViC and add data to meet their own needs without any prompting from the formal CIViC team. Finally, we have recruited domain experts and partnered with key organizations such as the ClinGen Somatic Working Group to moderate information in the database. These individuals typically provide targeted curation for specific genes, variants or diseases in the database. Although we have not yet performed a deep assessment of the reasons why curators are so active within the CIViC database, we hope that it is because it supports their own research and they believe in our mission to create a publically accessible and open-access database that contains high quality variant information to inform precision oncology.
What improvements are in the pipeline for CIViC? CIViC is on track for major upgrades to our server, client, and knowledgebase schema. For v2 we aim to provide a graph query language API to the knowledgebase on the server side, and a completely new user-interface that will emphasize community collaboration and knowledge discovery on the client side. An interim version 1.5 is planned to showcase an OpenAPI-compliant REST API, and an expanded knowledgebase schema. The v2 knowledgebase schema will include support for functional evidence, large-scale and complex variants with genotypes, and integration with clinical end-users.
Want to read more about CIViC? Check out the manuscript describing CIViC published in Nature Genetics. CIViC is open source and its github repository can be found at: https://github.com/griffithlab/civic-client and https://github.com/griffithlab/civic-server.
Special thanks to Erica Barnell, Obi Griffith, and Malachi Griffith for answering our questions about CIViC.
